Results
Simulations of protein diffusion and aggregation for bioprocess design and optimization
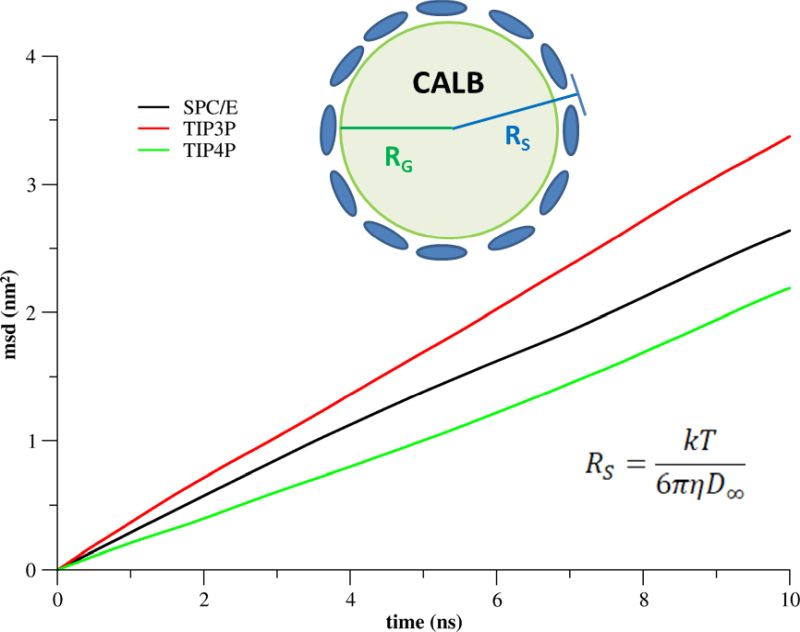
A single C.antarctica lipase B (CALB) molecule in water was simulated considering three different water models (SPC/E, TIP3P, TIP4P) by a series of MD simulations of three different box sizes (L = 9, 14, and 19 nm) to determine the diffusion coefficient, the water viscosity, and the effective protein size. The protein–water systems were equilibrated for 500 ns, followed by 100 ns production runs, which were subsequently analyzed. The diffusion of CALB was characterized by the Stokes radius (RS), which was derived from the diffusion coefficient and the water viscosity. RS was compared to the geometric radius (RG) of CALB, which was derived from the protein density. RS and RG differed by 0.27 nm for SPC/E and by 0.40 and 0.39 nm for TIP3P and TIP4P, respectively, which characterizes the thickness of the diffusive hydration layer on the protein surface (Fig. 3).
 |
|
The simulated hydration layer of CALB was in agreement with experimental data for seven different proteins of comparable size. Thus, the diffusion of proteins can be reliably simulated, if common pitfalls are avoided: (1) Different box sizes are simulated to account for the finite size effect. (2) The protein–water system is equilibrated sufficiently. (3) The complete production run is used for the determination of the diffusion coefficient.
Improving the productivity of a bioprocess requires detailed understanding of the behavior of the biocatalyst under process conditions, because high substrate concentrations, organic solvents, or elevated temperature promote protein aggregation. Molecular dynamics simulations were applied to study diffusion and aggregation of CALB in pure water and at high substrate concentration. In solution, substrate, additives, and solvent molecules compete for binding at the protein surface and thus modify the protein interactions and the aggregation behavior. Since the simulation of biocatalysts under process conditions provides insights into kinetic pathways of protein association as well as the molecular structure of aggregates, it is a valuable starting point for process engineering and enzyme design.
For more details:
Ferrario V, Pleiss J, 2018. Simulation of protein diffusion: a sensitive probe of protein-solvent interactions. J Biomol Struct Dyn (in press)
Kulschewski T, Pleiss J, 2013. A molecular dynamics study of liquid aliphatic alcohols: simulation of density and self-diffusion coefficient using a modified OPLS force field. Mol Simulat 39: 754 -767
Hutt M, Kulschewski T, Pleiss J, 2012. Molecular modelling of the mass density of single proteins. J Biomol Struct Dyn 30: 318 -327
Interpretation of kinetic modeling by thermodynamic activity
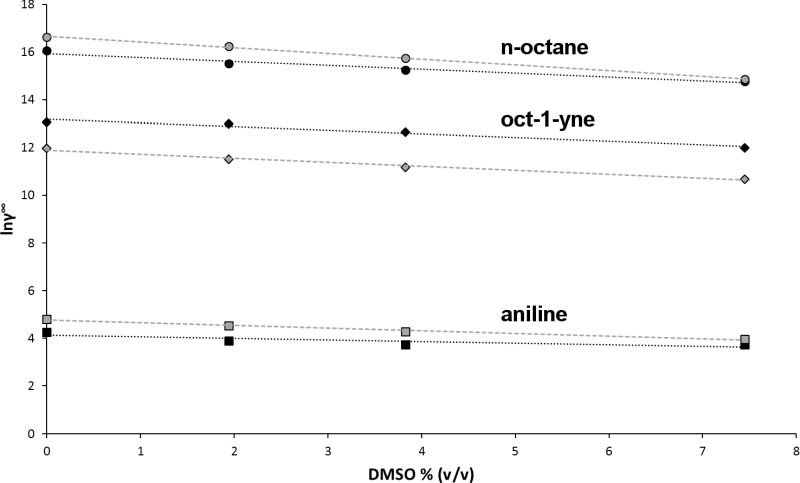
The experimentally determined Michaelis constant Kmc results from a combination of two effects: the recognition of the substrate by the enzyme and the molecular interactions between substrate and solvent. By separating substrate recognition from solvent effects, the thermodynamic activity-based Michaelis constant Kma allows for an unambiguous comparison of how different substrates fit into the substrate binding site. Kma of a poorly water-soluble substrate is calculated from the experimentally determined concentration-based Michaelis constant Kmc and its activity coefficient at infinite dilution γ∞. Comparing the Kma of different substrates instead of the experimentally determined Kmc prevents misinterpretations of molecular recognition. While n-octane showed the lowest Kmc value of six substrates of the cytochrome P450 monooxygenase P450BM-3, its Kma was 500 fold higher than aniline, indicating that the binding of n-octane is mainly driven by its low water solubility, while binding of aniline is driven by its shape complementarity. For three substrates (aniline, oct-1-yne, n-octane), γ∞ was reliably calculated by molecular dynamics simulations, either in binary substrate-water mixtures or in ternary mixtures including the co-solvent DMSO. It was demonstrated that the widely used co-solvent DMSO has a considerable effect on the measured Kmc value. Depending on the substrate, addition of 10% v/v DMSO increases Kmc by up to a factor of 11 (Fig. 4).
 |
|
To make biocatalytic experiments reproducible, it is therefore of utmost importance to carefully report the reaction conditions. The reliable simulation of activity coefficients in complex mixtures allows for an unambiguous comparison of enzyme-substrate interactions and provides a predictive tool for the design of biocatalytic processes.
For more details:
Ferrario V, Hansen N, Pleiss J, 2018. Interpretation of cytochrome P450 monooxygenase kinetics by modeling of thermodynamic activity. J Inorg Biochem (in press)
Pleiss J, 2018. Thermodynamic activity-based progress curve analysis in enzyme kinetics. Trends Biotechnol 36: 234 -238
Pleiss J, 2017. Thermodynamic activity–based interpretation of enzyme kinetics. Trends Biotechnol 35: 379 -382
Understanding enzyme-substrate interactions
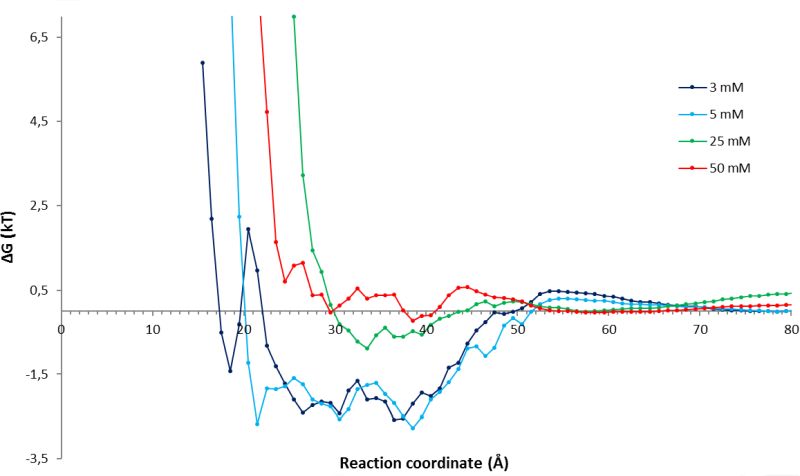
MD simulations were applied to analyze the interactions of substrate molecules with the enzyme in different solvent environment and at different concentrations. From long-time-scale simulations, the number of substrate molecules at a given distance from the active site was counted and interpreted as a free energy potential (Fig. 5).
 |
|
Understanding how the substrate diffuses from the protein surface to the enzyme active site opens the possibility to combine kinetic modelling of enzymes with a thermodynamic interpretation. Experimentally determined binding constants can be modeled at a molecular level, thus resulting in a deeper understanding enzyme function. The integration of kinetic, thermodynamic, and molecular modelling opens a new and promising route for rational enzyme and process engineering.
For more details:
Kulschewski T, Pleiss J, 2016. Binding of solvent molecules to a protein surface in binary mixtures follows a competitive Langmuir model. Langmuir 32: 8960 -8968
Sasso F, Kulschewski T, Secundo F, Lotti M, Pleiss J, 2015. The effect of thermodynamic properties of solvent mixtures explains the difference between methanol and ethanol in C.antarctica lipase B catalyzed alcoholysis. J Biotechnol 214: 1 -8
Kulschewski T, Sasso F, Secundo F, Lotti M, Pleiss J, 2013. Molecular mechanism of deactivation of C. antarctica lipase B by methanol. J Biotechnol 168: 462 -469

