Results
|
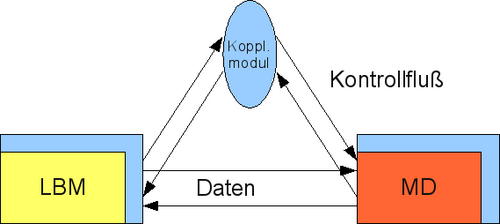
With ls1/Mardyn a parallel C++ molecular dynamics simulation code ls1/Mardyn, written by this working group, is available, which is enhanced in its functionality within project A.1 and optimized within this project. Project A.2 provides a Fortran90 Lattice Boltzmann Code and develops the theoretical basis for the coupling between a molecular dynamic and a continuum mechanical simulation approach to build a bridge between the nanoscopic and mesoscopic scale. The partitioned approach for the coupling of two independent, concurrent simulation programs necessitates an independent coupling module which handles the exchange of data and control.
A modern numerical simulation offers online visualization and provides possibilities for the user to intervene during run time. Especially molecular dynamics simulations have long run times on many processors and thus need many resources. It is imperative with regard to the later practical use in industrial development, to have control of the simulation run, as well as the possibility to correct simulation parameters during the simulation run.
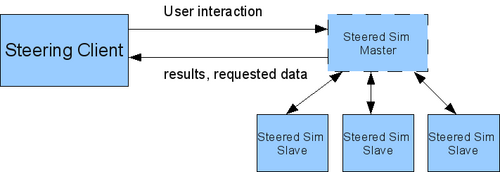
For these tasks this subproject develops and uses the computational steering framework Steereo. This framework, which is written using C++, offers steering modules to be used in simulations as well as in client applications, which then can request, process and change data from the simulation. At the moment, sequential and parallel (see figure 1) simulations written in C/C++ or Fortran90 (using a wrapper) are supported.
|
The framework itself is designed to be flexible: it is possible to program and integrate one's own modules to adapt to a given problem. That way, individual modules can be defined, which can be called from a client application. One can influence the simulation beyond mere parameter changes by doing that.
Besides a communication through POSIX TCP sockets, for parallel applications MPI communication will also be supported between these applications. For the coupling, a separate coupling module is developed, which controls and synchronizes the connection between the Lattice Boltzmann code from project A.2 with the molecular dynamics simulation from project A.1 (see figure 2).
|
For online visualization the visualization framework MegaMol from project D.3 is used, for which a Steereo using module has been written to request relevant data from the simulation during runtime.
- Description
- Publications
- Frontpage of this subproject