Biochemistry and Biophysics
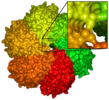
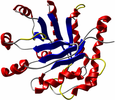 Biopolymers such as proteins or DNA are described as electrically charged macromolecules with a complex and well-defined structure, but also with a high degree of local flexibility. They form structures and aggregate into functional complexes. The balance of structure and flexibility and the high degree of specific, anisotropic interactions result in the interesting properties of biopolymers. In addition, the solvent plays a central role by mediating biophysical and biochemical processes. The simulation of DNA and proteins is based on complex, dynamic systems and requires the analysis of long time scales and large particle numbers.
Biopolymers such as proteins or DNA are described as electrically charged macromolecules with a complex and well-defined structure, but also with a high degree of local flexibility. They form structures and aggregate into functional complexes. The balance of structure and flexibility and the high degree of specific, anisotropic interactions result in the interesting properties of biopolymers. In addition, the solvent plays a central role by mediating biophysical and biochemical processes. The simulation of DNA and proteins is based on complex, dynamic systems and requires the analysis of long time scales and large particle numbers.
Molecular dynamics simulations for modelling of proteins and DNA
The method of molecular dynamics simulations is an important tool for the simulation of charged biopolymers. Typical biomolecular systems consist of 100000 atoms and are limited to a total simulation time of several 100 ns. However, the system size or the time scale that can be investigated at atomic resolution is not sufficient for applications such as protein folding or translocation of DNA which take place in a time scale of milliseconds or seconds. To model these processes, coarse-graining methods are applied.
In the SFB 716 methods are developed and applied to investigate biopolymers (proteins and DNA) in complex environments such as organic solvents, protein solutions, and solid surfaces, and to investigate complex processes such as protein-protein- and protein-ligand binding, and transport through pores.
Subprojects in this project area

